
Computational Biology is an important part of Coronavirus research. There are large, established distributed computing projects specifically working on COVID-19 research: Folding@home and Rosetta@home are two of the oldest and best-established projects. But both of those projects have not historically been made available to run on mobile devices with Arm processors.
We are actively working to provide Arm support for these Coronavirus distributed computing applications. This will make them available on the Neocortix Scalable Compute platform, and also on the BOINC platform running on Arm devices, and on other Arm-based platforms such as Raspberry Pi, MiniNodes and Packet.com.
Rosetta@home
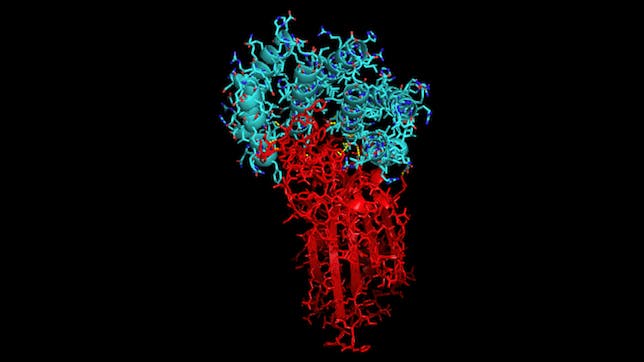
With the recent COVID-19 outbreak, Rosetta@home has been used to predict the structure of proteins important to the disease as well as to produce new, stable mini-proteins to be used as potential therapeutics and diagnostics, like the one displayed below which is bound to part of the COVID-19 spike protein:
From March 17-31, 2020, Neocortix worked with teams from Rosetta@home, Arm, MiniNodes, Linaro, and Packet.com to create, verify and test a Linux-on-Arm build of Rosetta. On March 31, the Rosetta team published the new Linux-on-Arm build.
Since then, the group effort has shifted to deployment onto Arm devices. On April 8, Neocortix completed its upgrades for temperature and battery monitoring, and is now ramping up deployment onto its existing network of 3000 mobile devices, soon to be followed by up to 42,000 volunteer devices from our PhonePaycheck waiting list.
As of April 16, the broader group has over 793 Arm devices contributing to the Rosetta project, with over 6.6M total credits earned. By May 6, Neocortix had ramped up to 613 simultaneous devices, and reached the Top 1% of Rosetta contributors with 2,303,633 total credits earned and a Recent Average Credit score of 102,963. By May 28, Neocortix has earned a total of 4,315,994 credits. By June 8, the broader group has 5628 devices contributing to Rosetta, with over 102.7M total credits earned.

Folding@home
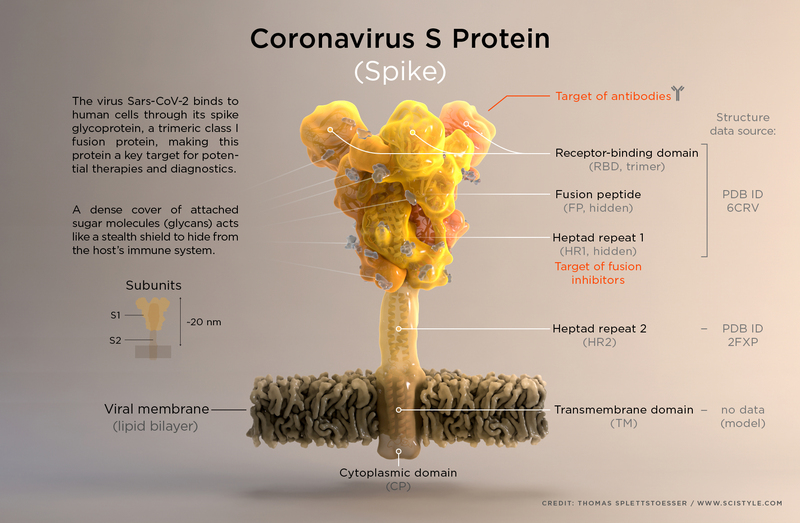
The spike of the SARS-CoV-2 virus (shown below) is a major target for designing therapeutics to combat the COVID-19 disease.
On April 1, 2020, the Folding@home team began its engagement with our extended team to create a Linux-on-Arm build of Folding@home. Neocortix immediately completed a Linux-on-Arm build of Gromacs, which is an important component of Folding@home.
On April 16, Neocortix completed a build of FaHCore with the new Gromacs 2020.1 engine, for both x86_64 and aarch64 targets, with SIMD optimization (AVX2_256 and ARM_NEON_ASIMD, respectively). On April 20, Neocortix completed a build of FaHClient. On April 24, Neocortix provided a performance-improved version of FaHCore using MPI, OpenMP, and Arm Performance Libraries. On May 5, the Neocortix, Gromacs and Folding@home teams completed a number of outstanding tasks to allow Folding@home to fully use the Gromacs 2020.1 engine. On May 20, the changes were accepted and merged into the Folding@home master branch.
The next step was to make Folding@home Core builds for all supported platforms: Windows, Linux, macOS, with CUDA. On May 26, the Folding@home team asked for help to complete those builds, and Neocortix agreed to do it. On May 30, Neocortix completed the builds, and the Folding@home team accepted and internally published the cores. On May 31, the Folding@home team established an internal project for testing the new A8 version of the Core with real Work Units. Testing is currently under way. On June 7, the Folding@home team requested more variations of CUDA/Intel builds, and Dmitry provided them. Through the month of June, the extended teams identified and solved several problems in the F@h Core. On July 2, Dmitry solved the last remaining problem with resuming a terminated job from a checkpoint, and submitted the F@h Core version A8 0.0.4 to the Folding@home team.
By July 7, the extended team had reached a consensus that F@h Core version A8 0.0.4 was ready to ship. Dr. Bowman called a meeting for July 13 to discuss release planning. On July 8, Dmitry and Joseph got the Folding@home servers reconfigured to give real work units to Arm devices, and shortly thereafter, Dmitry got Folding@home running on a real work unit in a Neocortix Scalable Compute instance on his Samsung Galaxy S20 phone in his lab.
On July 10, Michael got Folding@home running on real work units on Neocortix Scalable Compute instances on 10 phones on our worldwide network of PhonePaycheck devices. On July 12, the first work units completed. By August 6, we had completed 3,000 work units.

On July 13, the teams from Neocortix, Arm, Folding@home and Gromacs met to plan for publication and announcements for the Linux-on-Arm version of Folding@home. We agreed that we would aim for July 28, to allow two weeks for Folding@home to do more beta testing and develop device-specific job sizing. On July 28, Neocortix launched its press release, as planned. But unfortunately, Folding@home did not publish their Linux-on-Arm version at that time.
On August 10, Folding@home published a
beta version of the Linux-on-Arm release.
On October 28, Folding@home published an
official version of the Linux-on-Arm release.
And on November 24, Folding@home
officially announced the Linux-on-Arm release.

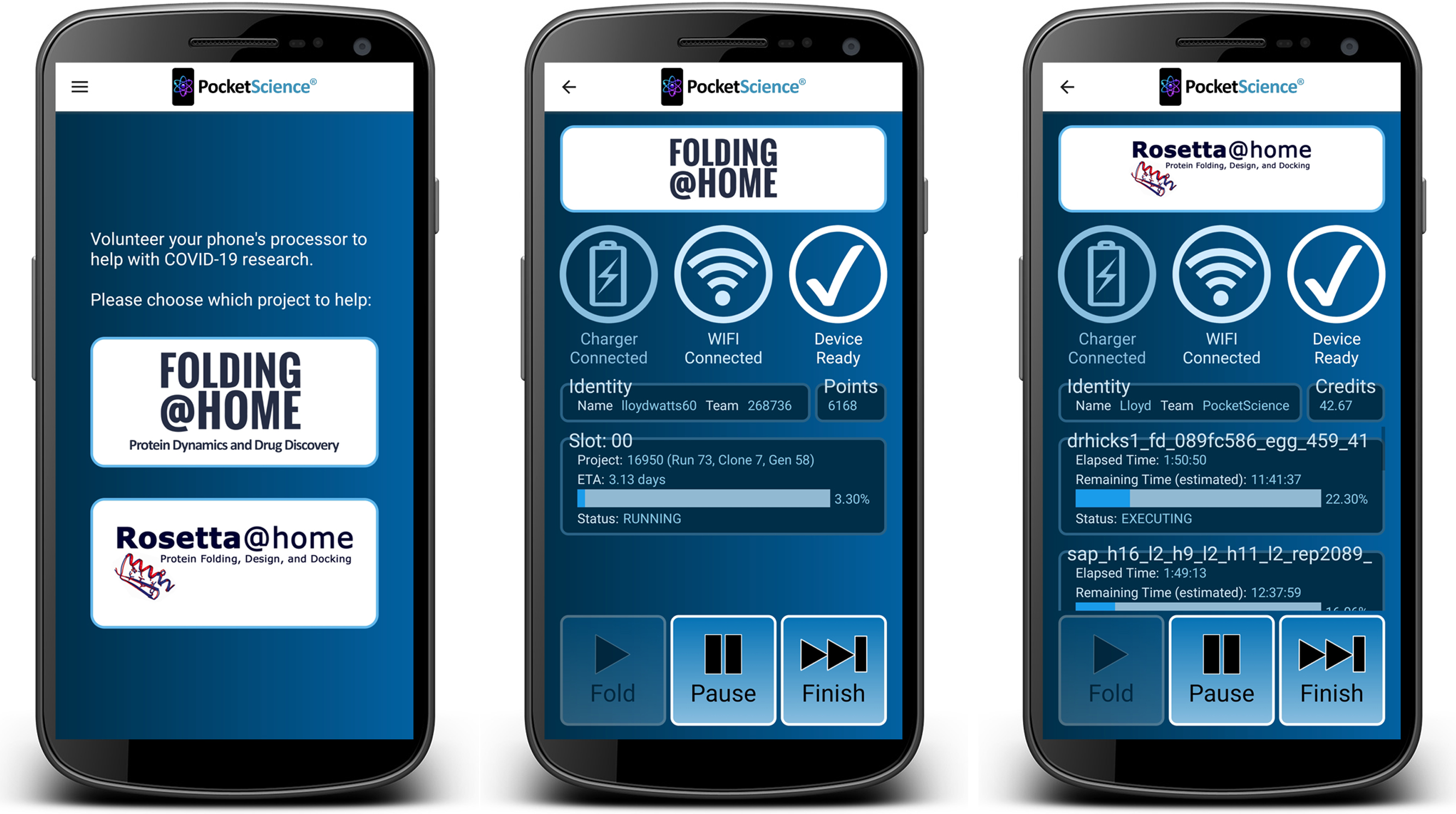
On February 22, 2021, our PocketScience Android app was published to the
Google Play Store, allowing potentially millions of mobile phones to contribute
to COVID-19 research using the Folding@home and Rosetta@home distributed computing applications.
Videos
On February 26, 2022, we removed our PocketScience Android app from the Google Play Store. Since Coronavirus vaccines are now widely available, there is much less interest in contributing computing power to Coronavirus research.
Acknowledgments
We are grateful for support and collaboration from the following companies and organizations: